Advancing Pediatric and Longitudinal DNA Methylation Studies with CellsPickMe, an Integrated Blood Cell Deconvolution Method
Advancing Pediatric and Longitudinal DNA Methylation Studies with CellsPickMe, an Integrated Blood Cell Deconvolution Method
Fu, M. P.; Edwards, K.; Navarro-Delgado, E. I.; Merrill, S. M.; Kitaba, N. T.; Konwar, C.; Mandhane, P.; Simons, E.; Subbarao, P.; Moraes, T. J.; Holloway, J. W.; Turvey, S. E.; Kobor, M. S.
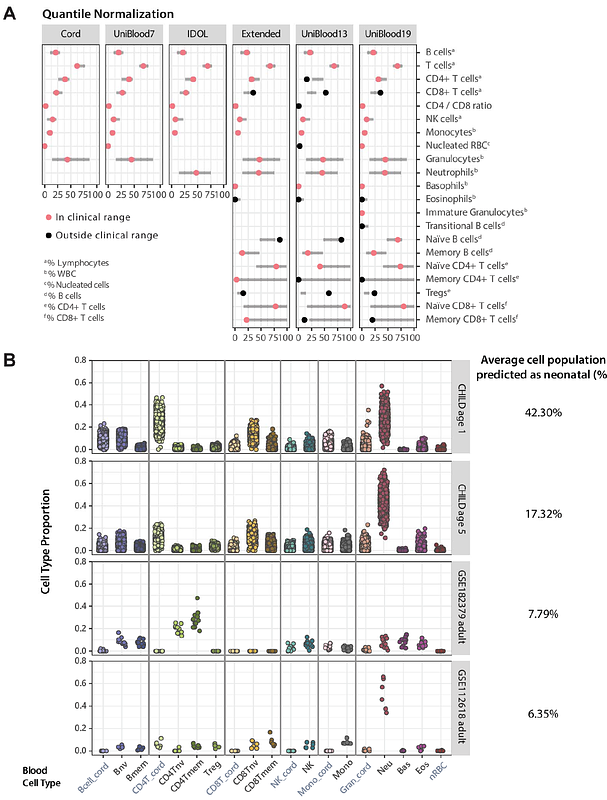
AbstractProspective birth cohorts offer the potential to interrogate the relation between early life environment and embedded biological processes such as DNA methylation (DNAme). These association studies are frequently conducted in the context of blood, a heterogeneous tissue composed of diverse cell types. Accounting for this cellular heterogeneity across samples is essential, as it is a main contributor to inter-individual DNAme variation. Integrated blood cell deconvolution of pediatric and longitudinal birth cohorts poses a major challenge, as existing methods fail to account for the distinct cell population shift between birth and adolescence. In this paper, we critically evaluated the reference-based deconvolution procedure and optimized its prediction accuracy for longitudinal birth cohorts using DNAme data from the Canadian Healthy Infant Longitudinal Development (CHILD) cohort. The optimized algorithm, CellsPickMe, integrates cord and adult references and picks DNAme features for each population of cells with machine learning algorithms. It demonstrated improved deconvolution accuracy in cord, pediatric, and adult blood samples compared to existing benchmark methods. CellsPickMe supports blood cell deconvolution across early developmental periods under a single framework, enabling cross-time-point integration of longitudinal DNAme studies. Given the increased resolution of cell populations predicted by CellsPickMe, this R package empowers researchers to explore immune system dynamics using DNAme data in population studies across the life course.