Structure from Noise: Confirmation Bias in Particle Picking in Structural Biology
Structure from Noise: Confirmation Bias in Particle Picking in Structural Biology
Balanov, A.; Zabatani, A.; Bendory, T.
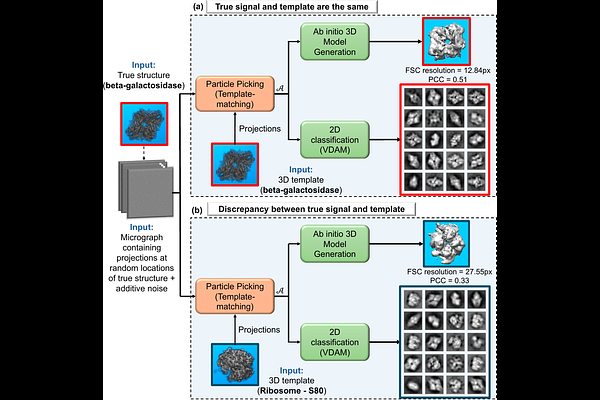
AbstractConfirmation bias is a fundamental challenge in cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET), where prior expectations can lead to systematic errors in data interpretation. This bias may emerge at multiple stages of the reconstruction pipeline, and in particular in the critical particle-picking stage, where 2D particles (in cryo-EM) or 3D subtomograms (in cryo-ET) are extracted from highly noisy micrographs or tomograms. Focusing on two widely used methodologies, template matching and deep neural networks, we combine theoretical analysis with controlled experiments to demonstrate that both methods, when applied to pure noise, can produce persistent molecular structures, a phenomenon we term structure from noise. This artifact highlights a critical vulnerability in current workflows: the potential for particle-picking algorithms to inject strong prior-driven bias into downstream analyses. We then propose practical mitigation strategies to reduce the impact of such biases. Together, our findings deepen the theoretical understanding of confirmation bias in cryo-EM and cryo-ET and call for cautious interpretation of reconstructions, primarily when relying on template-driven particle picking.