Fast, flexible gene cluster family delineation with IGUA
Fast, flexible gene cluster family delineation with IGUA
Larralde, M.; Blom, J.; Gourle, H.; Carroll, L. M.; Zeller, G. M.
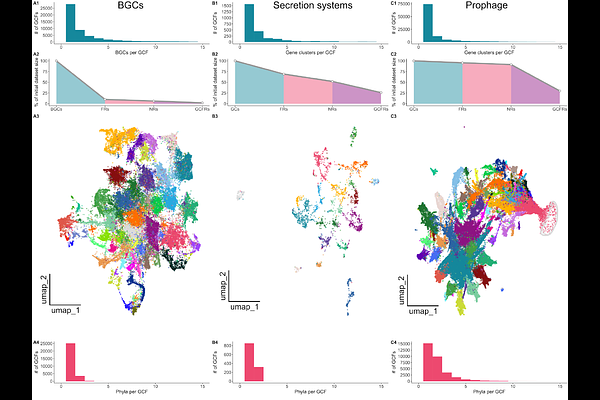
AbstractProkaryotic genomes harbor a variety of functional elements encoded as contiguous multi-gene clusters, with biosynthetic gene clusters (BGCs, genetic determinants of secondary metabolite biosynthesis) serving as a notable example. In a typical workflow, BGCs are clustered into Gene Cluster Families (GCFs), units that group BGCs encoding similar biosynthetic pathways together. However, existing methods cannot readily scale to massive datasets and cannot be used for GCF delineation tasks beyond BGC clustering. Here, we present IGUA (Iterative Gene clUster Analysis; https://github.com/zellerlab/IGUA), a scalable, flexible GCF delineation method for genomic segments with multi-gene architectures. On a BGC clustering task, IGUA is [≥]10x faster than the state-of-the-art (BiG-SCAPE/BiG-SLiCE), without sacrificing accuracy. To highlight its scalability, we use IGUA to cluster >2.8 million BGCs from {approx}1 million prokaryotic genomes in <18 hours (n = 2,829,071 BGCs to 56,960 GCFs). To showcase its utility beyond BGC clustering, we use IGUA to cluster (i) secretion systems and (ii) prophages into GCFs (n = 10,576 and 356,776 gene clusters to 2,744 and 213,699 GCFs, respectively). Overall, IGUA represents a versatile GCF delineation tool with unmatched computational efficiency and flexibility, enabling (meta)genomic mining applications at unprecedented scales.