Uncovering Functional Gene Regulatory Networks in Bulk and Single-Cell Data through Robust Transcription Factor Activity Estimation and Model-Guided Experimental Validation
Uncovering Functional Gene Regulatory Networks in Bulk and Single-Cell Data through Robust Transcription Factor Activity Estimation and Model-Guided Experimental Validation
Siahpirani, A. F.; McCalla, S. G.; Pyne, S.; Dillingham, C. M.; Sridharan, R.; Roy, S.
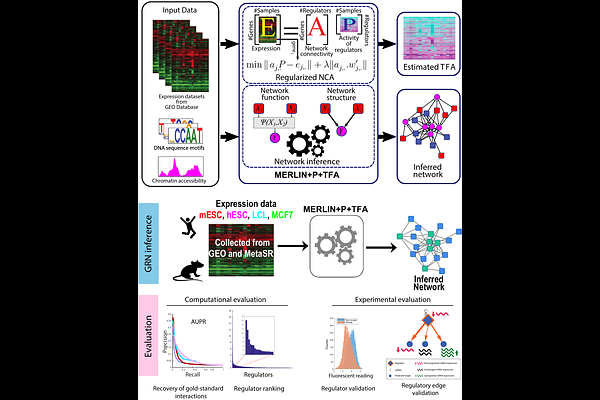
AbstractReconstructing genome-scale gene regulatory networks (GRNs) remains a difficult problem in systems biology, and many experimental and computational methods have been developed to address this problem. Recent computational methods have aimed to more accurately model GRNs by estimating the hidden Transcription Factor Activity (TFA), from prior knowledge of TF target regulatory connections, encoded as an input directed graph, to relax the assumption that mRNA level of the regulator correlates with the protein activity of the regulator. However, the noise in the prior knowledge can adversely affect the estimated TFA levels and the quality of the downstream inferred GRNs. Here, we present a new approach, MERLIN+P+TFA, that uses prior knowledge-guided sparsity regularization to robustly and accurately estimate TFA and downstream GRNs. We apply our method to simulated and real expression data in yeast and mammalian systems and show improved quality of inferred GRNs for both bulk and single-cell datasets. Regularized TFA offers benefits to a variety of other GRN inference algorithms, including those that have traditionally be used with expression alone, in both bulk and scRNA-seq settings. We used the inferred GRN to prioritize key regulators for the mouse Embryonic Stem Cell (mESC) state and validate 58 regulators experimentally. We identify both known and novel regulators of the mESC state and further validate the targets of 4 known and novel regulators. Our validation experiments suggest that computationally inferred networks can capture functional targets of TFs with higher precision than estimated in current benchmarks, however, it is important to generate context-specific gold standards.