Exapted CRISPR-Cas12f homologs drive RNA-guided transcription
Exapted CRISPR-Cas12f homologs drive RNA-guided transcription
Hoffmann, F. T.; Wiegand, T.; Palmieri, A. I.; Glass-Klaiber, J.; Xiao, R.; Tang, S.; Le, H.; Meers, C.; Lampe, G. D.; Chang, L.; Sternberg, S. H.
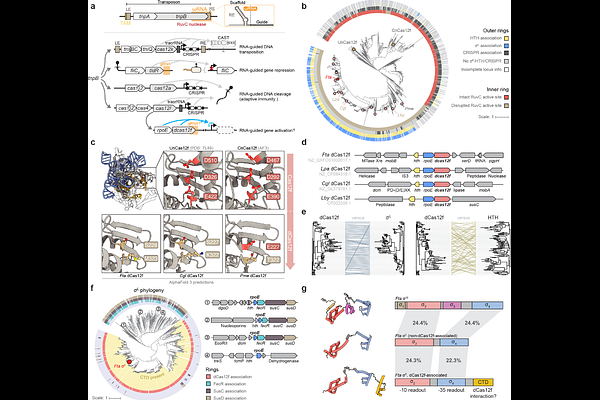
AbstractBacterial transcription initiation is a tightly regulated process that canonically relies on sequence-specific promoter recognition by dedicated sigma ({sigma}) factors, leading to functional DNA engagement by RNA polymerase (RNAP)(1). Although the seven {sigma} factors in E. coli have been extensively characterized(2), Bacteroidetes species encode dozens of specialized, extracytoplasmic function {sigma} factors ({sigma}E) whose precise roles are unknown, pointing to additional layers of regulatory potential(3). Here we uncover an unprecedented mechanism of RNA-guided gene activation involving the coordinated action of {sigma}E factor in complex with nuclease-dead Cas12f (dCas12f). We screened a large set of genetically-linked dCas12f and {sigma}E homologs in E. coli using RIP-seq and ChIP-seq experiments, revealing systems that exhibited robust guide RNA enrichment and DNA target binding with a minimal 5\'-G target-adjacent motif (TAM). Recruitment of {sigma}E was dependent on dCas12f and guide RNA (gRNA), suggesting direct protein-protein interactions, and co-expression experiments demonstrated that the dCas12f-gRNA-{sigma}E ternary complex was competent for programmable recruitment of the RNAP holoenzyme. Remarkably, dCas12f-RNA-{sigma}E complexes drove potent gene expression in the absence of any requisite promoter motifs, with de novo transcription start sites defined exclusively by the relative distance from the dCas12f-mediated R-loop. Our findings highlight a new paradigm of RNA-guided transcription (RGT) that embodies natural features reminiscent of CRISPRa technology developed by humans(4,5).