Decoding PDI diversity: insights into structure, domains, and functionality in sorghum
Decoding PDI diversity: insights into structure, domains, and functionality in sorghum
Lopez Gomez, C. F.; Morris, M. T.; Massel, K.; Smith, M.; Crisp, P.; Schenk, G.; Godwin, I. D.
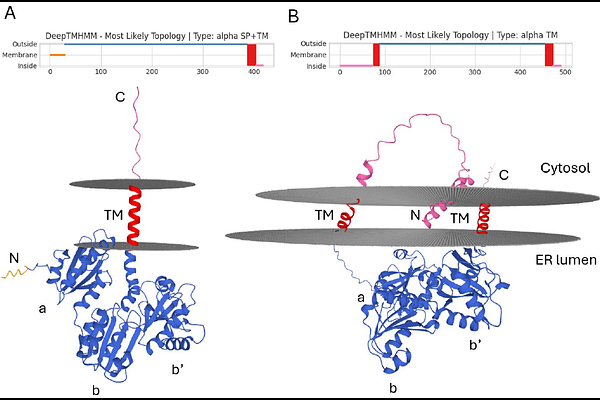
AbstractProteins play indispensable roles in cellular function, acting as both structural components and catalysts for essential biological processes. Their proper folding into three-dimensional structures is critical for functionality. To ensure correct folding, proteins interact with chaperones and folding catalysts such as Protein Disulfide Isomerases (PDIs), which assist in the formation and rearrangement of disulfide bonds that stabilize proteins by linking cysteine residues. PDIs are part of the thioredoxin (TRX) superfamily and are characterized by a conserved CXXC motif that contributes to their redox potential. They exhibit isomerase and oxidoreductase activities, that enable them to rearrange and form new disulfide bonds. PDI family members in sorghum (SbPDI) present a broad and largely unexplored diversity in domain order, structure, and architecture between or even within species. To shed light on this diversity, we identified and characterized PDI family members in sorghum in silico to explore their domain architecture, three-dimensional structure and functionality.