SARS-CoV-2 reshapes mA methylation in long non-coding RNAs of human lung cells
SARS-CoV-2 reshapes mA methylation in long non-coding RNAs of human lung cells
Peter, C. M.; Cyrino, C. O.; Moretti, N. R.; Antoneli, F.; Briones, M. R.
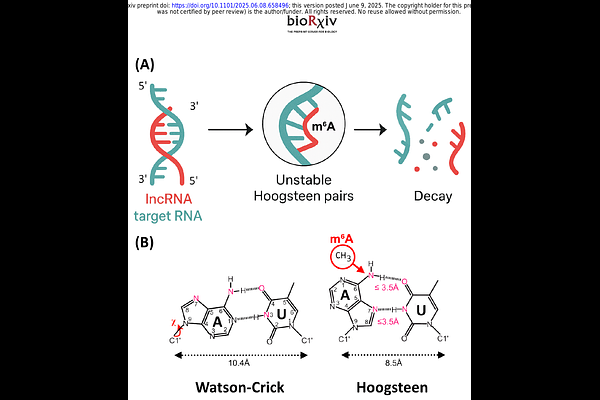
AbstractN6-Methyladenosine (m6A) is a key base modification that regulates RNA stability and translation during viral infection. While m6A methylation of host mRNAs has been studied in SARS-CoV-2-infected cells, its role in long non-coding RNAs (lncRNAs) is unknown. Here, we analyzed direct RNA sequencing (dRNA-seq) data from infected human lung cells (Calu-3) using a machine learning m6A detection framework. We observed a global increase in m6A levels across ten antiviral response-associated lncRNAs, with UCA1, GAS5, and NORAD--regulators of interferon (IFN) signaling--showing the most pronounced changes. This might, in part, explain the attenuated IFN expression observed in infected cells. We identified methylated DRACH motifs in predicted lncRNA duplex-forming regions, which may favor Hoogsteen base-pairing, which destabilize secondary structures and target interaction sites. These results provide new perspectives on how SARS-CoV-2 could impact lncRNAs to modulate host immunity and viral persistence through m6A-dependent mechanisms.