A fast TMT-based proteomic workflow reveals neural enrichment in neurospheres of hiPSC-derived neural stem cells
A fast TMT-based proteomic workflow reveals neural enrichment in neurospheres of hiPSC-derived neural stem cells
Muniz, P. d. L.; Souza, L. R. Q.; Martins, M. R.; Valadares, W.; Nogueira, F. C. S.; Rehen, S.; Nugue, G.; Junqueira, M.
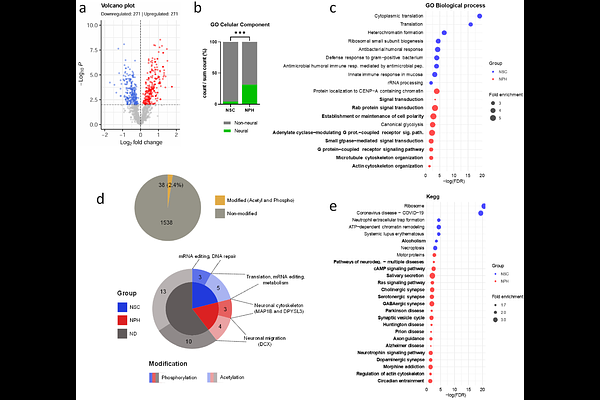
AbstractThree-dimensional (3D) neural spheroids, or neurospheres, generated from human induced pluripotent stem cell (hiPSC)-derived neural stem cells (NSCs) more accurately recapitulate the microenvironmental cues of neural tissue compared to traditional two-dimensional (2D) monolayers. However, comparative omics-based characterizations of these models remain limited. Here, we present a streamlined and scalable TMT-based quantitative proteomics workflow to contrast the proteomic landscapes of hiPSC-derived NSCs cultured in 2D monolayers versus 3D neurospheres. A total of 1,576 proteins were identified in an unfractionated LC-MS/MS of 68 minutes, with 542 showing significant differential abundance between groups. Neurospheres exhibited enrichment in neural-related pathways, such as synaptic signaling, neurotrophin signaling, cytoskeletal organization and vesicle trafficking, while monolayers enriched for multipotency features, such as general metabolic activity. Cell-type enrichment analyses confirmed increased neuronal identity in neurospheres, including elevated levels of markers associated with neuronal maturation. Our results demonstrate that 3D culture of NSCs induces a proteomic shift toward a more mature neural phenotype. This rapid, multiplexed proteomic approach enables high-content molecular profiling suitable for drug screening and personalized medicine applications.