H3K4me3 is a post-transcriptional histone mark
H3K4me3 is a post-transcriptional histone mark
Walvekar, K. P.; Chilaka, S.
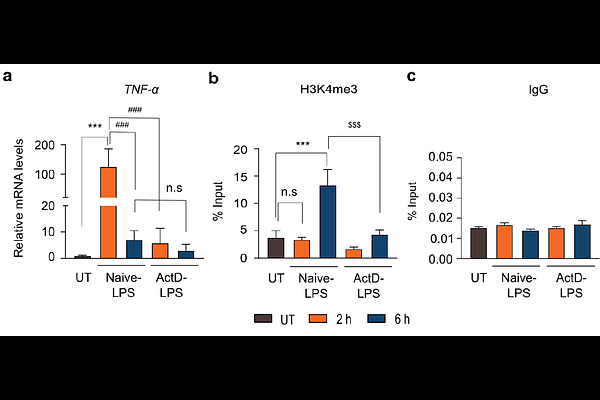
AbstractHistone H3 Lysine 4 trimethylation (H3K4me3) is widely recognized as a hallmark of actively transcribed gene promoters, yet its precise role in transcriptional regulation remains unresolved1-4. Here we present the first direct evidence that H3K4me3 is not a cause but a consequence of transcriptional activation, functioning as a downstream epigenetic response. Using inducible gene models to study temporally resolved transcriptional dynamics, we demonstrate that H3K4me3 enrichment significantly lags behind the transcriptional peaks of inflammatory genes such as TNF- and IL-1{beta}, appearing hours after maximal RNA synthesis. This delayed deposition contrasts sharply with the early accumulation of other active histone marks, including H3K27ac and H4K8ac, and with the recruitment of canonical transcriptional regulators such as phosphorylated RNA polymerase II, NF-{kappa}B, and chromatin modifier, histone acetyltransferase p300. Pharmacological inhibition of transcription with Actinomycin D reduces H3K4me3 enrichment, while knockdown of MLL1 abolishes H3K4me3 without affecting gene activation, indicating that H3K4me3 deposition is transcription-dependent but dispensable for transcriptional initiation. Furthermore, analysis of the constitutively active MYC gene suggests that post-transcriptional H3K4me3 deposition may be a general feature of active transcription. Together, these findings redefine H3K4me3 as a post-transcriptional histone mark, revealing an unexpected layer of epigenetic regulation with broad implications for gene expression control.