TrAEL-seq captures DNA replication dynamics in mammalian cells
TrAEL-seq captures DNA replication dynamics in mammalian cells
Kara, N.; Biggins, L.; Grinkevich, V.; Whale, A.; Garran Garcia, P.; Srinivasan, J.; Rugg-Gunn, P.; Andrews, S.; Parry, A.; Robinson, H. M. R.; Houseley, J.
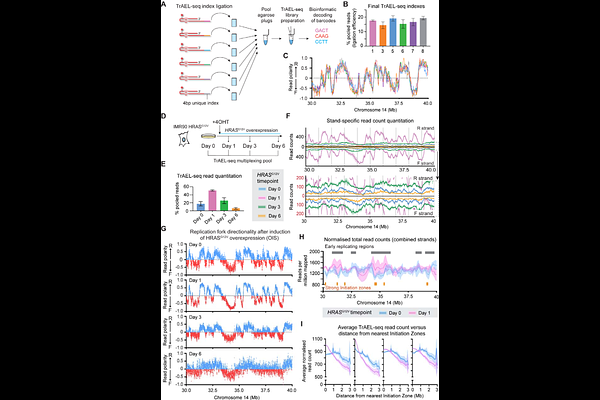
AbstractPrecise DNA replication is critical to the maintenance of genome stability, and the DNA replication machinery is a focal point of many current and upcoming chemotherapeutics. TrAEL-seq is a robust method for profiling DNA replication genome-wide that works in unsynchronised cells and does not require treatment with drugs or nucleotide analogues. Here, we provide an updated method for TrAEL-seq including multiplexing of up to 6 samples that dramatically improves sample quality and throughput, and we validate TrAEL-seq in multiple mammalian cell lines. The updated protocol is straightforward and robust yet provides excellent resolution comparable to OK-seq in mammalian cell samples. High resolution replication profiles can be obtained across large panels of samples and in dynamic systems, for example during the progressive onset of oncogene induced senescence. In addition to mapping zones where replication initiates and terminates, TrAEL-seq is sensitive to replication fork speed, revealing effects of both transcription and proximity to replication Initiation Zones on fork progression. Although forks move more slowly through transcribed regions, this does not have a significant impact on the broader dynamics of replication fork progression, which is dominated by rapid fork movement in long replication regions (>1Mb). Short and long replication regions are not intrinsically different, and instead replication forks accelerate across the first ~1 Mb of travel such that forks progress faster in the middle of regions lying between widely spaced Initiation Zones. We propose that this is a natural consequence of fewer replication forks being active later in S phase when these distal regions are replicated and there being less competition for replication factors.