DIA-PASEF Proteomic Profiling Reveals MpkA-Dependent Iron Stress Responses and Siderophore Biosynthesis in Aspergillus nidulans
DIA-PASEF Proteomic Profiling Reveals MpkA-Dependent Iron Stress Responses and Siderophore Biosynthesis in Aspergillus nidulans
Lee, J.; West, O. K.; Huso, W.; Doan, A. G.; Gray, K. J.; Edwards, H.; Tran, J. T.; Carman, D. R.; Betenbaugh, M.; Srivastava, R.; Harris, S.; Marten, M. R.
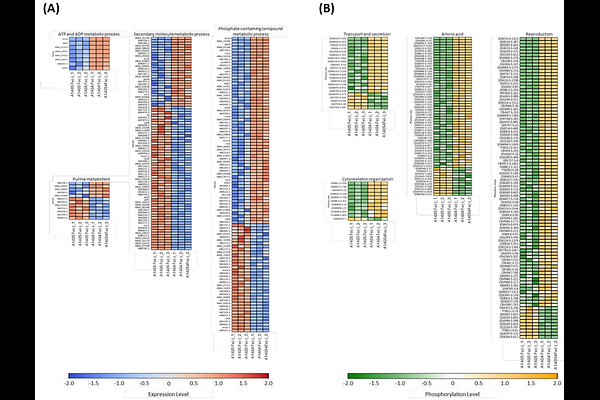
AbstractFilamentous fungi play essential roles in biotechnology as producers of valuable bioproducts and, conversely, as opportunistic pathogens. Aspergillus nidulans is a widely used model organism for fungal genetics and cell biology; however, comprehensive proteomic references for this species remain limited. In this study, we applied a data-independent acquisition parallel accumulation serial fragmentation (DIA-PASEF) approach to enable efficient and in-depth proteome profiling of A. nidulans. Leveraging ion mobility-based ion cloud information from DDA-PASEF experiments, we developed DIA-PASEF methods that identified 3,904 proteins across biological triplicates grown in rich medium. Compared to prior studies, this represents an increase in protein identifications of more than 140% and was achieved with more than five-fold reduction in analysis time. We employed this newly developed DIA&ndash] PASEF methodology to conduct both proteomic and phosphoproteomic analyses under iron-depleted conditions in an MpkA protein-kinase deficient mutant ({Delta}mpkA). The {Delta}mpkA strain exhibited expression of approximately 500 additional proteins and occupancy of over 1,800 additional phosphosites relative to a control. Differentially expressed and phosphorylated proteins increased by more than an order of magnitude in the {Delta}mpkA mutant across both iron-replete and iron- deplete conditions. Gene Ontology (GO) enrichment analysis revealed broader and distinct biological processes under iron-depleted conditions, highlighting adaptive responses specific to iron limitation and MAPK pathway disruption. This work establishes a high-coverage proteomic resource for A. nidulans and provides novel insights into fungal stress responses and signaling network perturbation. Importantly, high-throughput proteomic profiling reveals that limited iron availability and MAPK pathway disruption increases siderophore biosynthesis.