SSAlign: Ultrafast and Sensitive Protein Structure Search at Scale
SSAlign: Ultrafast and Sensitive Protein Structure Search at Scale
Wang, L.; Zhang, X.; Wang, Y.; Xue, Z.
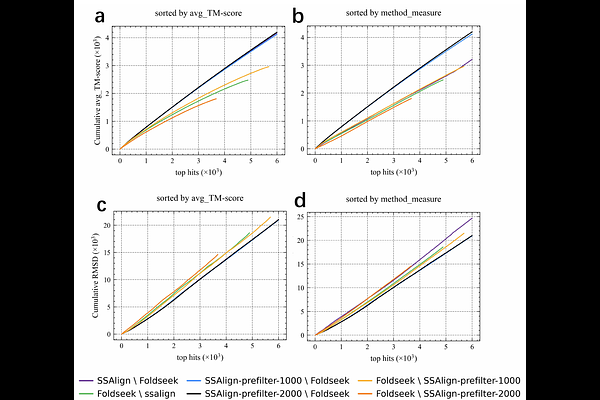
AbstractThe advent of highly accurate structure prediction techniques such as AlphaFold3 is driving an unprecedented expansion of protein structure databases. This rapid growth creates an urgent demand for novel search tools, as even the current fastest available methods like Foldseek face significant limitations in sensitivity and scalability when confronted with these massive repositories. To meet this challenge, we have developed SSAlign, a protein structure retrieval tool that leverages protein language models to jointly encode sequence and structural information, and adopts a two-stage alignment strategy optimized with multi-GPU and multi-process parallelization. On large-scale datasets such as AFDB50, SSAlign outpaces Foldseek by two to three orders of magnitude in search speed, offering unmatched scalability for high-throughput structural analysis. Compared to Foldseek, SSAlign retrieves substantially more high-quality matches on Swiss-Prot and achieves marked performance improvements on SCOPe40, with relative AUC increases of +20.2% at the family level and +33.3% at the superfamily level, demonstrating significantly enhanced sensitivity and recall. In sum, SSAlign achieves TM-align-comparable accuracy with Foldseek-surpassing speed and coverage, offering an efficient, sensitive, and scalable solution for large-scale structural biology and structure-based drug discovery.