Machine Learning-Driven Drug Repurposing for KRAS G12C and KRAS G12D Inhibition
Machine Learning-Driven Drug Repurposing for KRAS G12C and KRAS G12D Inhibition
Fuschi, G.; Germain, J. S.; Bebensee, D.; Moawad, C.; Aladysheva, A.; Mohamed, A.; Elwakeel, E.; Brooks, B. R.; Amin, M.
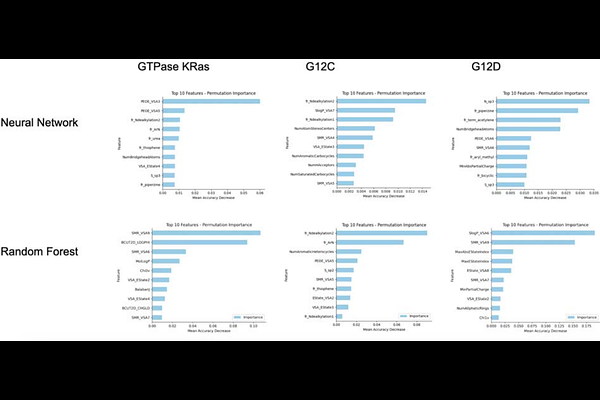
AbstractKRAS is a predominant oncogenic driver across multiple cancers, long considered undruggable due to its high nucleotide affinity and lack of classical binding pockets. Although recent advances have led to covalent inhibitors like Sotorasib and Adagrasib for the KRAS G12C mutation, effective therapies for other common variants most notably KRAS G12D, which is highly prevalent in aggressive pancreatic cancers remain limited. In this study, we employ machine learning to identify potential inhibitors for both KRAS G12D and G12C by screening FDA-approved compounds from the ChEMBL database. Random Forest and Neural Network models were trained on binding affinity data from three BindingDB datasets: wild-type KRAS GTPase, KRAS G12C, and KRAS G12D. Our models identified high-affinity candidates including anti-cancer kinase inhibitors (e.g., Cobimetinib, Gilteritinib) as well as drugs from other categories (e.g., Bromocriptine, Cefepime). By incorporating atomic hybridization as a feature, we aim to capture the effects of induced polarization, potentially improving prediction accuracy. Despite modest agreement across models, our approach highlights several promising candidates such as Acalabrutinib, which has prior evidence of G12C activity for further investigation. These results offer a data-driven foundation for experimental validation and the future development of targeted KRAS therapies.