Structural motif search across the protein-universe with Folddisco
Structural motif search across the protein-universe with Folddisco
Kim, H.; Kim, R. S.; Mirdita, M.; Steinegger, M.
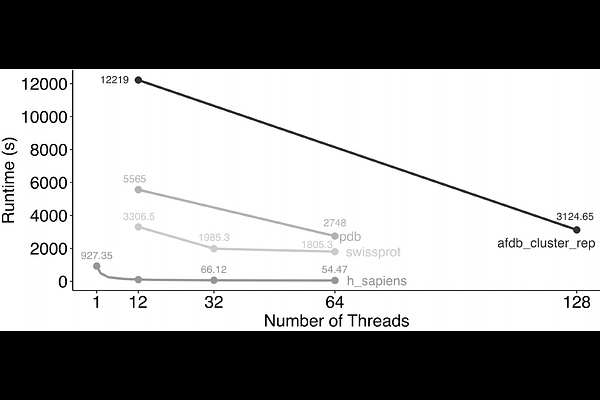
AbstractDetecting similar protein structural motifs, functionally crucial short 3D patterns, in large structure collections is computationally prohibitive. Therefore, we developed Folddisco, which overcomes this through an index of position-independent geometric features, including side-chain orientation, combined with a rarity-based scoring system. Folddisco indexes 53 million AFDB50 structures into 1.45 terabyte within 24 hours, enabling rapid detection of discontinuous or segment motifs. Folddisco is more accurate and storage-efficient than state-of-the-art methods, while being an order of magnitude faster. Folddisco is free software available at folddisco.foldseek.com and a webserver at https://search.foldseek.com/folddisco.