Designing Novel Solenoid Proteins with In Silico Evolution
Designing Novel Solenoid Proteins with In Silico Evolution
Pretorius, D.; Nikov, G. I.; Washio, K.; Florent, S.-W.; Taunt, H.; Ovchinnikov, S.; Murray, J. W.
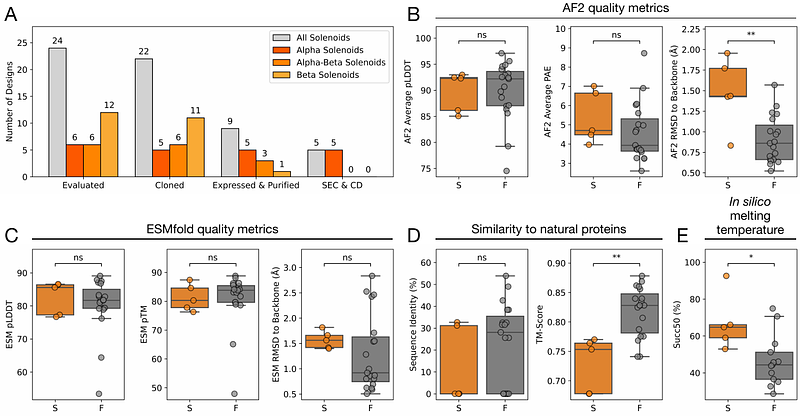
AbstractSolenoid proteins are elongated tandem repeat proteins with diverse biological functions, making them attractive targets for protein design. Advances in machine learning have transformed our understanding of sequence-structure relationships, enabling new approaches for de novo protein design. Here, we present an in silico evolution platform that couples a solenoid discriminator network with AlphaFold2 as an oracle within a genetic algorithm. Starting from random sequences, we design -, {beta}-, and {beta}-solenoid backbones, generating structures that span natural and novel solenoid space. We experimentally characterise 41 solenoid designs, with -solenoids consistently folding as intended, including one structurally validated design that closely matches the design model. All {beta}-solenoids initially failed, reflecting the difficulty of designing {beta}-strand majority proteins. By introducing terminal capping elements and refining designs based on earlier experimental screens, we generate two {beta}-solenoids that have biophysical properties consistent with their designs. Our approach achieves fold-specific hallucination-based design without depending on explicit structural templates.