Engineering microbial consortia for distributed signal processing
Engineering microbial consortia for distributed signal processing
Duncker, K. E.; Shende, A. R.; Shyti, I.; Ruan, A.; D'Cunha, R.; Ma, H. R.; Venugopal Lavanya, H.; Liu, S.; Gottel, N.; Anderson, D. J.; Gunsch, C. K.; You, L.
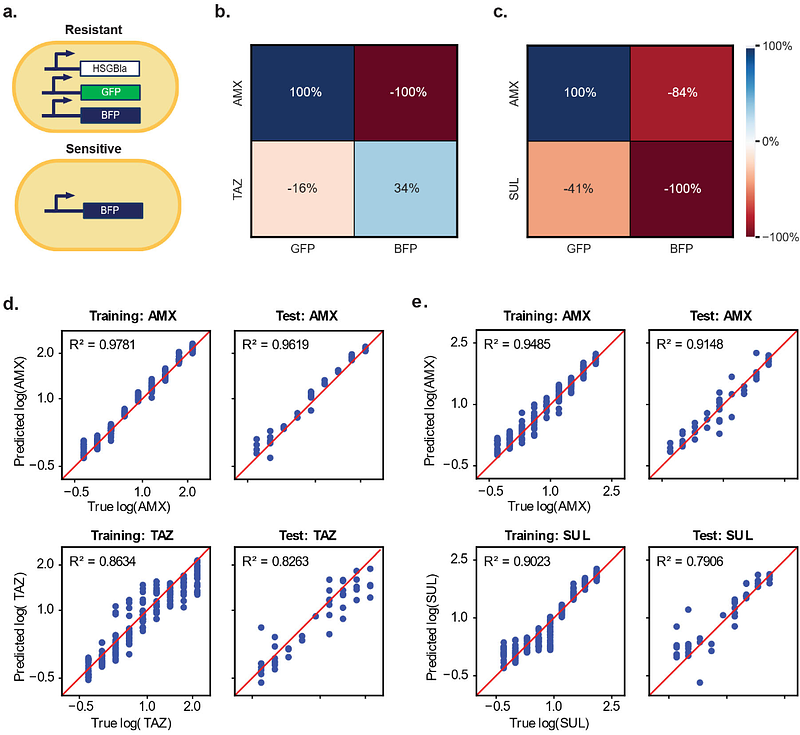
AbstractA critical goal in biology is deducing input signals from measurable readouts. Genetic circuits have successfully been designed to respond to specific analytes and produce quantifiable outputs; however, multiplexed signal processing remains challenging. This limitation is partially due to crosstalk, or sensors\' non-specific responses to unintended signals. While strategies to achieve orthogonality are promising, they are time-intensive and context-dependent. Here, we introduce a new, generalizable approach that leverages microbial consortia to distribute sensory functions and computational methods to disentangle signals. Compartmentalizing sensor circuits within distinct populations simplifies experimental optimization by allowing individual populations to be exchanged without requiring genetic modifications. Our computational pipeline combines mechanistic modeling with machine learning to decode microbial communities\' unique temporal responses and predict multiple input concentrations. We demonstrated this platform\'s versatility in a variety of contexts: measuring signals with high crosstalk, detecting antibiotics with natural microbial communities, and quantifying chemicals in hospital sink wastewater. Our findings highlight how combining microbial engineering with computational strategies can produce robust, scalable biosensors for diverse applications.