{Psi}-Atlas: an integrated atlas for pseudouridine epitranscriptome
{Psi}-Atlas: an integrated atlas for pseudouridine epitranscriptome
Wang, X.; Luo, J.; Lang, X.; Liu, Q.
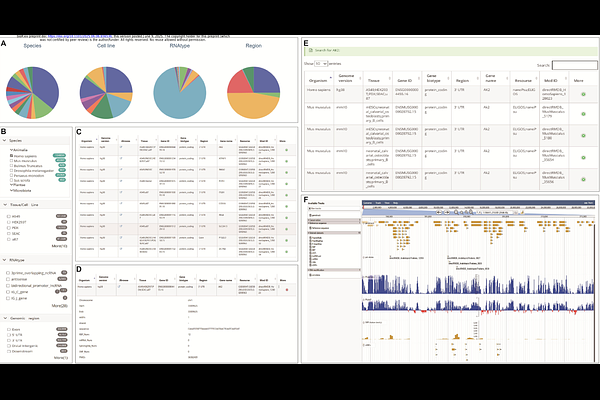
AbstractPseudouridine ({Psi}) is a C5 glycoside isomer of uridine, formed by breaking the N1 glycosyl bond and undergoing a 180{degrees} base rotation. This modification stands as one of the most widespread post-transcriptional alterations in RNA and is universally distributed among diverse RNA species. The pervasiveness of this modification enhances RNA structural integrity, bestows unique structural and functional attributes upon the RNA molecules it adorns, and facilitates additional hydrogen bonding. However, the absence of a convenient and integrated intuitive visualization database encompassing all currently reported species and RNA types is evident. Here, we present the {Psi}-Atlas, an extensive database meticulously curated for the comprehensive collection and annotation of RNA pseudouridine. This database encompasses 554,895 pseudouridine modification sites across various RNA categories, including mRNA, ncRNA, tRNA, rRNA, and others, in 55 distinct species reported in current literature. The sequencing methodologies employed comprise the next-generation sequencing (NGS) techniques such as {Psi}-Seq, Pseudo-Seq, CeU-Seq, PSI-Seq, RBS-Seq, HydraPsiSeq, BID-Seq, PRAISE-Seq, as well as third-generation sequencing methods like Direct RNA sequencing (DRS). {Psi}-Atlas as the most comprehensive and integrated resource for RNA pseudouridine modifications to date. And the {Psi}-Atlas database offers an intuitive interface for information display and a myriad of analytical tools, including PsiVar and PsiFinder. In essence, this platform serves as a robust search and visualization tool for the study of pseudouridylation in epitranscriptomics. {Psi}-Atlas is available at https://rnainformatics.org.cn/PsiAtlas.