Visual-like Template Diffusion: Boosting Single-Sequence Protein Structure Prediction by Adapting Image Diffusion Models
Visual-like Template Diffusion: Boosting Single-Sequence Protein Structure Prediction by Adapting Image Diffusion Models
Wang, X.; Zhang, T.; Cui, Z.; Guo, X.; Wang, F.; Wang, Y.; Cai, X.; Zheng, W.
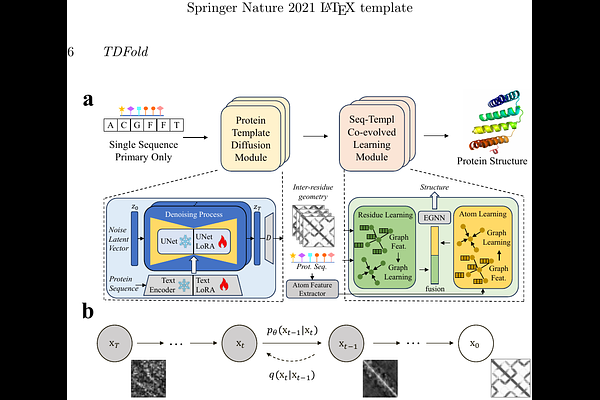
AbstractSingle-sequence protein structure prediction has drawn increasing attention due to the high computational costs associated with obtaining homologous information. Here, we propose a visual-like template diffusion method, named TDFold, to achieve accurate and highly efficient single-sequence 3D structure prediction for proteins. Given a protein sequence, TDFold initially generates high-quality inter-residue geometries (distances and orientations) as templates from a probabilistic diffusion perspective. Since inter-residue geometries can be encoded as multi-channel feature matrices (each channel for either the inter-residue distance or orientation correlation), analogous to image features, we construct an image-level template diffusion module by adapting the stable diffusion (SD) model from text-vision generation to sequence-template diffusion for proteins. Subsequently, a lightweight sequence-template co-evolved learning (SCL) network is constructed to facilitate accurate and efficient protein structure prediction. As a result, TDFold possesses three highlights: (i) better single-sequence prediction performance: TDFold greatly outperforms existing protein language models (PLMs), including ESMFold (also known as ESM2), OmegaFold, trRosettaX-Single, and RGN2, on homology-insufficient datasets such as Orphan and Orphan25, while also achieving promising results on the popular CASP14 and CASP15 benchmarks; (ii) low resource consumption: By utilizing the lightweight SCL architecture, the GPU memory consumption of TDFold is generally lower than that of popular methods such as AlphaFold2 and ESMFold; (iii) higher efficiency in training and inference: TDFold can be trained within a week using a single NVIDIA 4090 GPU. Furthermore, the inference time of TDFold is significantly shorter (about 10x to 100x) than that of existing methods (ESMFold and AlphaFold2) for long protein sequences. This work demonstrates the effectiveness of leveraging powerful vision diffusion models to enhance protein template generation, thereby establishing a new paradigm for single-sequence protein structure prediction. It also accelerates protein-related research, particularly for resource-limited universities and academic institutions. The code has been released to speed up biological research.