Human protein interactome structure prediction at scale with Boltz-2
Human protein interactome structure prediction at scale with Boltz-2
Ille, A. M.; Markosian, C.; Burley, S. K.; Pasqualini, R.; Arap, W.
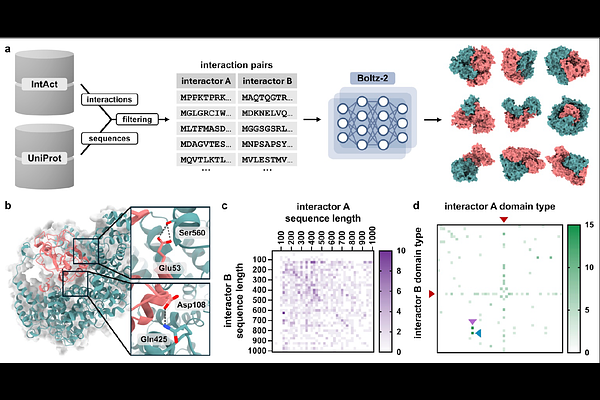
AbstractIn humans, protein-protein interactions mediate numerous biological processes and are central to both normal physiology and disease. Extensive research efforts have aimed to elucidate the human protein interactome, and comprehensive databases now catalog these interactions at scale. However, structural coverage of the human protein interactome is limited and remains challenging to resolve through experimental methodology alone. Recent advances in artificial intelligence/machine learning (AI/ML)-based approaches for protein interaction structure prediction present opportunities for large-scale structural characterization of the human interactome. One such model, Boltz-2, which is capable of predicting the structures of protein complexes, may serve this objective. Here, we present de novo computed models of 1,394 binary human protein interaction structures predicted using Boltz-2. These structural predictions were based on biochemically determined interaction data sourced from the IntAct database, together with corresponding protein sequences obtained from UniProt. We then assessed the predicted interaction structures through various confidence metrics, which consider both overall structure and the interaction interface. These analyses indicated that prediction confidence tended to be greater for smaller complexes, while increased multiple sequence alignment (MSA) depth tended to improve prediction confidence. Additionally, we performed a limited evaluation against experimentally determined human protein complex structures not included in the Boltz-2 training regimen, which indicated prediction accuracy consistent with AlphaFold3. This work demonstrates the utility of Boltz-2 for in silico structural modeling of the human protein interactome, while highlighting both strengths and limitations. Ultimately, such modeling is expected to yield broad structural insights with relevance across multiple domains of biomedical research.