Ish: SIMD and GPU Accelerated Local and Semi-Global Alignment as a CLI Filtering Tool
Ish: SIMD and GPU Accelerated Local and Semi-Global Alignment as a CLI Filtering Tool
Stadick, S.
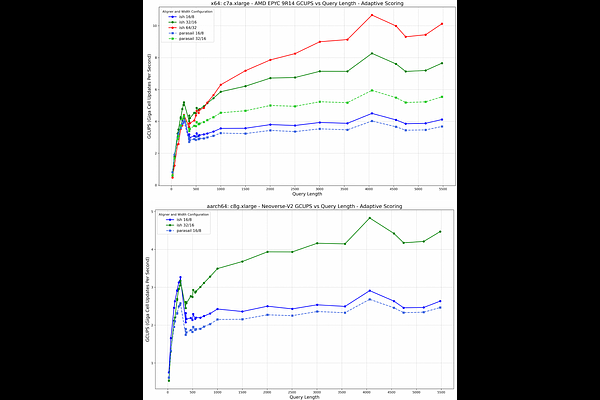
AbstractBackground: Filtering records using command line tools is a staple of Bioinformatics. In analysis pipelines and in day-to-day research tools such as awk, grep, and cut are the workhorses of much of our data crunching. To date, there is no command line utility for performing index-free alignment-based filtering of records. Results: Ish is a classic and composable unix-style CLI tool. It takes a query and filters the input target records to only those that match the input query with a threshold alignment score, using a selectable alignment algorithm. The core alignment algorithms for Ish meet or exceed the performance of their reference implementations for both SIMD and GPU alignment, as measured by gigacell updates per second (GCUPs). Conclusions: Ish fills a longstanding gap in Bioinformatics tooling by offering a CLI utility for index-free alignment-based record filtering. Additionally, it provides meaningfully improved versions of the classic striped local and semi-global full dynamic programming algorithms, along with a GPU-based semi-global alignment method.