Boosting AlphaFold Protein Tertiary Structure Prediction through MSA Engineering and Extensive Model Sampling and Ranking in CASP16
Boosting AlphaFold Protein Tertiary Structure Prediction through MSA Engineering and Extensive Model Sampling and Ranking in CASP16
Liu, J.; Neupane, P.; Cheng, J.
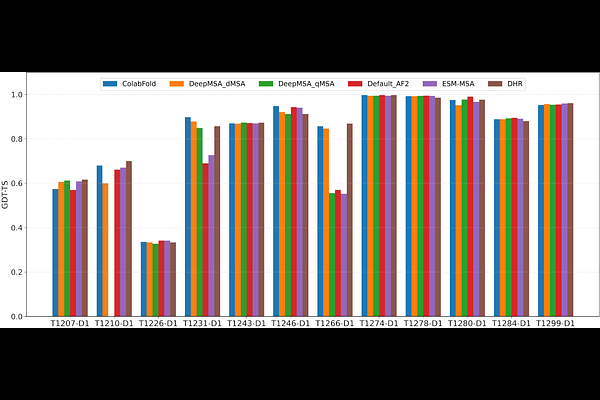
AbstractAlphaFold2 and AlphaFold3 have revolutionized protein structure prediction by enabling high-accuracy tertiary structure predictions for most single-chain proteins. However, obtaining high-quality predictions for some hard protein targets with shallow or noisy multiple sequence alignments (MSAs) and complicated multidomain architectures remains challenging. Here, we present MULTICOM4, an integrative protein structure prediction system that uses diverse MSA generation, large-scale model sampling, and an ensemble model quality assessment (QA) strategy of combining individual QA methods to improve model generation and ranking of AlphaFold2 and AlphaFold3. In the 16th Critical Assessment of Techniques for Protein Structure Prediction (CASP16), our predictors built on MULTICOM4 ranked among the top performers out of 120 predictors in tertiary structure prediction and outperformed a standard AlphaFold3 predictor. The average TM-score of our best performing predictor MULTCOM\'s top-1 prediction for 84 CASP16 domain is 0.902. It achieved high accuracy (TM-score > 0.9) for 73.8% of the 84 domains and correct fold predictions (TMscore > 0.5) for 97.6% domains in terms of top-1 prediction. In terms of best-of-top-5 prediction, it predicted correct folds for all the domains. The results show that MSA engineering through the use of different protein sequence databases, alignment tools, and domain segmentation as well as extensive model sampling are the key to generate accurate and correct structural models. Additionally, using multiple complementary QA methods and model clustering can improve the robustness and reliability of model ranking.