Riemann-GNN: Causal Reasoning on Hyperbolic Riemannian Manifolds for Interpretable Drug-Disease Prediction
Riemann-GNN: Causal Reasoning on Hyperbolic Riemannian Manifolds for Interpretable Drug-Disease Prediction
Yang, J.; Puyu, H.; Liu, Q.; Pan, Y.; He, L.; Zhang, L.; Dai, L.; Wang, Y.; Tao, J.
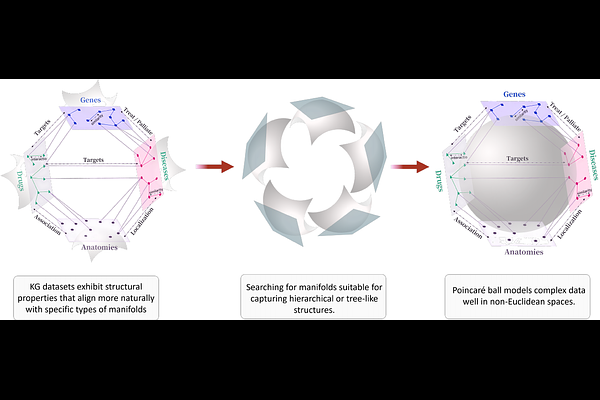
AbstractThe high cost and low success rate of de novo drug development have driven increasing interest in drug repurposing. Computational approaches to drug repurposing leverage large-scale biomedical knowledge graphs (KGs) to identify new indications for existing drugs. However, the structural heterogeneity, relation sparsity, and limited interpretability of current models remain significant challenges. In this study, we propose Riemann-GNN, an end-to-end framework that combines hyperbolic geometry, graph neural networks (GNNs), and causal inference for interpretable drug repurposing. Riemann-GNN embeds multi-relational biomedical KGs into a hyperbolic Riemannian manifold modeled by the Poincare ball, effectively capturing the hierarchical structure of biomedical entities. On top of this geometry, we introduce a causal reasoning module that extracts disease-centric subgraphs and applies intervention-based inference using structural causal models. This module provides mechanistic explanations for predicted drug-disease associations. We evaluate our model on PrimeKG, a comprehensive biomedical KG with over 129,000 nodes and 4 million edges, achieving AUROC = 0.971, AUPRC = 0.963, and Recall@20 = 0.847, significantly out performing several state-of-the-art baselines. In a case study on amyotrophic lateral sclerosis (ALS), Riemann-GNN identifies 20 novel candidate drugs--such as Bosutinib, Masitinib, and Tamoxifen--and provides interpretable causal paths explaining their therapeutic relevance. Several predictions without existing ALS labels in the KG are supported by literature evidence, while others suggest new hypotheses for future validation. These results demonstrate the utility of integrating hyperbolic representation learning with causal semantics for robust and explainable biomedical AI.