PIMENTO: A PrIMEr infereNce TOolkit to facilitate large-scale calling of amplicon sequence variants
PIMENTO: A PrIMEr infereNce TOolkit to facilitate large-scale calling of amplicon sequence variants
Atallah, C.; Richardson, L.; Beracochea, M.; Finn, R. D.
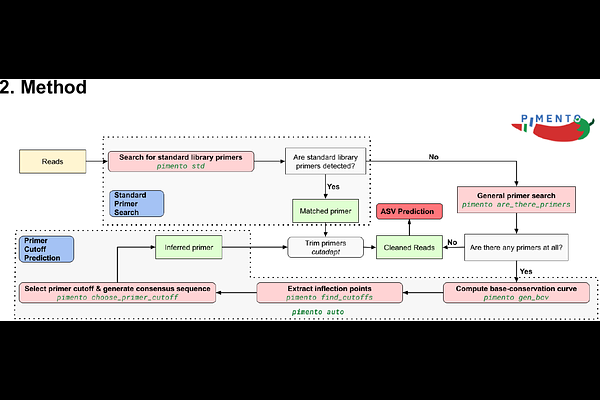
AbstractThe calling of amplicon sequence variants from DNA metabarcoding data is a common method of revealing the taxonomic makeup of environmental samples. A significant hurdle to the large-scale calling of amplicon sequence variants from publicly available nucleotide datasets is the presence of primer sequences in reads, the removal of which is a necessary pre-processing step for this form of analysis. Further, as the details of which primers were used is rarely associated with the sequence records, there is a need for a method that can automatically infer the presence and identity of primers in sequencing data. In this work, we introduce PIMENTO, a Python package which uses a dual-strategy approach for identifying primers that are present in sequencing reads to enable their removal, and therefore facilitate amplicon sequence variant calling at scale.