Rapid and accurate protein structure database search using inverse folding model and contrastive learning
Rapid and accurate protein structure database search using inverse folding model and contrastive learning
Lyu, Q.; Wei, H.; Chen, S.; Peng, Z.; Yang, J.
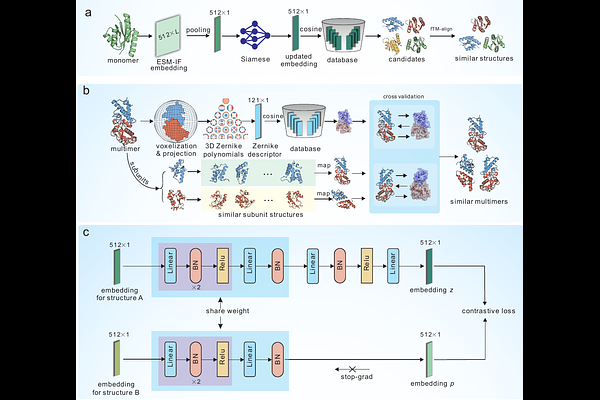
AbstractProtein structure database search has become increasingly challenging due to the growing number of experimental and computational structures. We introduce mTM-align2, a novel two-step approach for rapid and accurate protein structure database search. In the first step, protein structures are first transformed into embeddings using a pre-trained inverse folding model (ESM-IF) and 3D Zernike polynomials. The ESM-IF embeddings are further optimized through a contrastive learning network, which is trained on ~7 million structure pairs. Structures with similar embeddings are returned on the fly in this step. The second step employs a rapid structure alignment program to refine top candidates, ensuring high precision and producing high-quality alignments. Extensive benchmarks reveal that mTM-align2 performs competitively compared to other leading methods, completing monomeric structure search in seconds with over 90% precision for the top 10 hits. The t-SNE visualization of the mTM-align2 embeddings for thousands of structures demonstrates that our embeddings are structurally informed, capturing the global structural features. It uncovers insights such as structure misclassifications and ambiguous structural class boundaries. A web server for mTM-align2 is accessible at https://yanglab.qd.sdu.edu.cn/mTM-align/.