eRNAs Modulate mRNA Stability and Translation Efficiency to Bridge Transcriptional and Post-transcriptional Gene Regulation
eRNAs Modulate mRNA Stability and Translation Efficiency to Bridge Transcriptional and Post-transcriptional Gene Regulation
Kuklinkova, R.; Benova, N.; Anene, C. A.
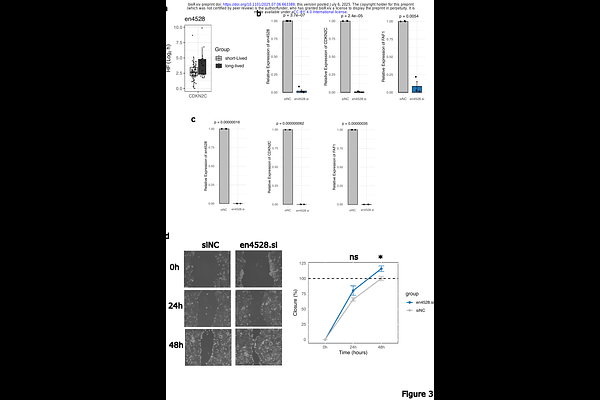
AbstractEnhancer RNAs (eRNAs) are best known for their role in transcriptional regulation, where they facilitate enhancer-promoter communication and chromatin remodelling. Yet growing evidence suggests that their function may extend beyond the nucleus. Here, we systematically characterise the decay kinetics of eRNAs across human cell types using time-resolved transcriptomics and kinetic modelling. While most eRNAs undergo canonical exponential decay, a subset displays non-linear dynamics, suggesting context-dependent degradation mechanisms. Perturbation of core decay regulators, including components of the mA and CCR4-NOT pathways, reveals that eRNA stability is modulated by a patchwork of pathways governing mRNA turnover. Integrating transcriptome-wide ribosome profiling, RNA-Seq, and half-life data, we identify eRNAs associated with changes in mRNA stability and translation efficiency of their target protein-coding transcripts. Functional validation of one such eRNA, en4528, shows it regulates CDKN2C mRNA independently of transcription and impacts cell migration. These findings redefine the regulatory scope of eRNAs, positioning them as active participants in post-transcriptional gene control and cellular behaviour. The resulting decay profiles and regulatory annotations have been incorporated into the eRNAkit database, available at https://github.com/AneneLab/eRNAkit, enhancing its capacity for integrative systems-level analysis of eRNA function.