TARRAGON: Therapeutic Target Applicability Ranking and Retrieval-Augmented Generation Over Networks
TARRAGON: Therapeutic Target Applicability Ranking and Retrieval-Augmented Generation Over Networks
Beasley, J.-M.; Schatz, K.; Ding, E.; DeLuca, M.; Abu Zaid, N.; Tucker, N.; Chirkova, R.; Crona, D.; Tropsha, A.; Muratov, E.
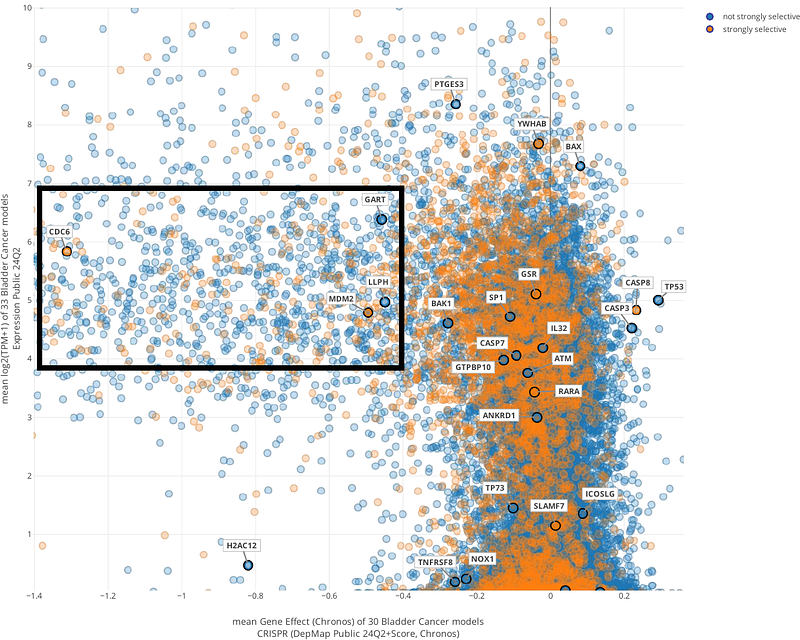
AbstractThe identification of therapeutic protein targets is fundamental to the success of drug development and repurposing. Traditional approaches for target selection require extensive preclinical evaluation for toxicity and efficacy, making the process time-intensive and resource-heavy. Computational tools that efficiently prioritize and validate novel targets are needed to streamline drug discovery workflows. To address this gap, we developed TARRAGON: Therapeutic Target Applicability Ranking and Retrieval-Augmented Generation Over Networks, a computational framework that integrates data mining and machine learning to identify, rank, and assess target-disease relationships to nominate new therapeutic targets. TARRAGON mines knowledge graphs to uncover meta-paths, or rules of graph traversal, linking potential therapeutic targets to diseases. It employs a classification model to rank target-disease hypotheses based on evidence patterns and utilizes a retrieval-augmented generation workflow to prompt a large language model for generating feasibility reports on prioritized targets. Using TARRAGON, we prioritized potential drug targets for non-muscle invasive urinary bladder cancer. Top-ranked candidates were validated using CRISPR gene effect and expression data from the Broad Institute DepMap portal. We further proposed chemical modulators for these targets to inform combination drug screening alongside approved bladder cancer therapeutics. TARRAGON introduces a novel, interpretable computational pipeline for therapeutic target discovery and pharmaceutical candidate nomination, offering the potential to accelerate drug development across diverse disease areas.